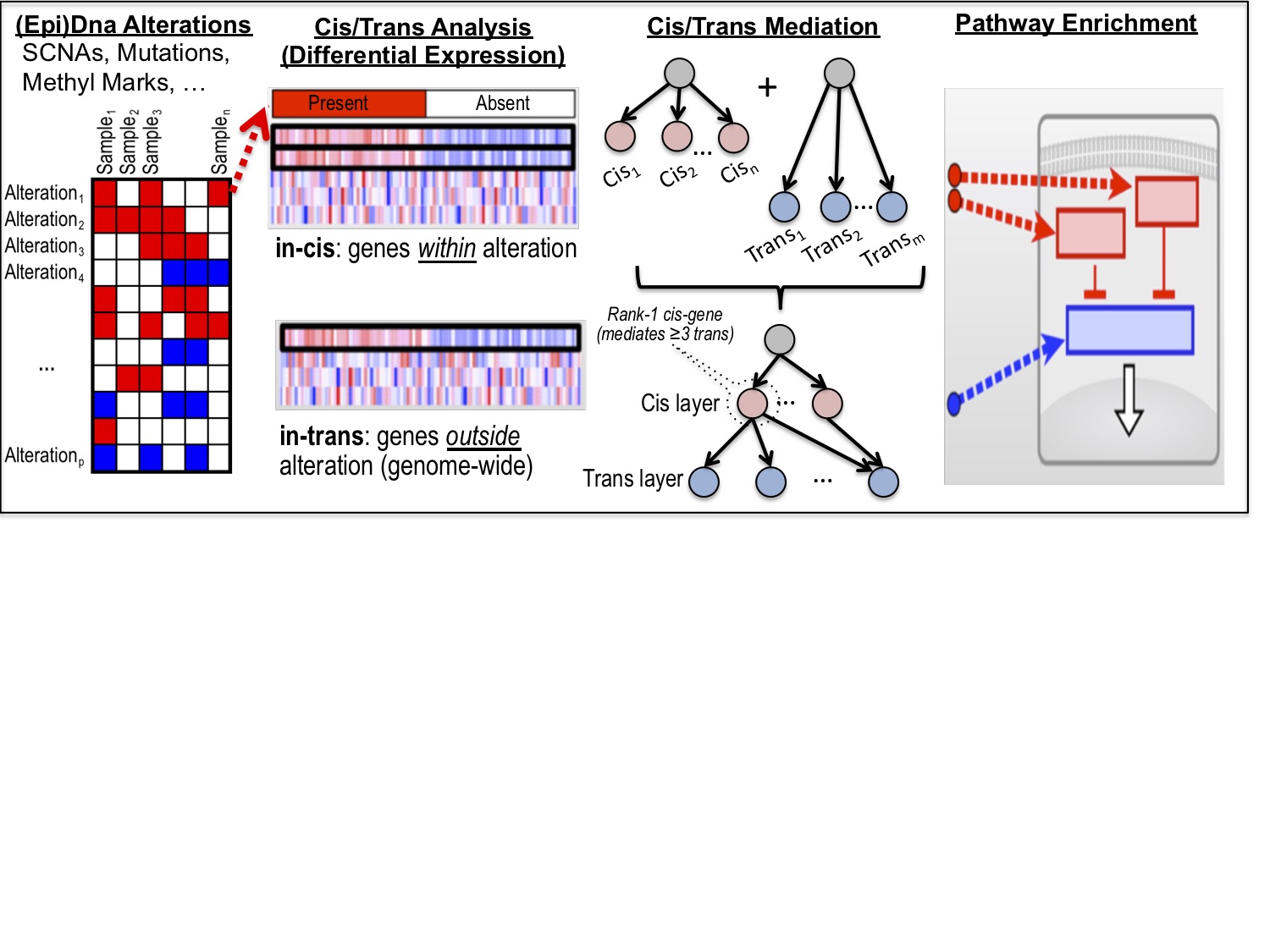
iEDGE is a computational tool for performing integrative analysis of epi-DNA and gene expression data. This tool was based on methods described in Monti, Chapuy, et. al. (Cancer Cell, 2012) for integrating copy number alteration with gene expression data for studying Diffuse Large B cell Lymphoma, and has since been extended to TCGA pan-cancer analysis of Somatic Copy Number (SCNA) and Gene Expression Data in 19 cancer types. The generalized pipeline performs integrative analysis of epi-genomic data (methylation, copy number alteration, mutation, microRNA, etc) and gene-expression data arising from overlapping samples and consists of 3 major modules:
Citation:
Here, we present the web portal reports from the TCGA pancancer analysis. Detailed reports in additional formats (.txt) are available upon request.